Mutation and Genetic Variation
Mutations are changes in the DNA sequence of an organism. They can have various causes, including errors during DNA replication or damage from environmental factors. Mutations are categorized based on their effect on the genetic material, and understanding them is crucial in molecular ecology for interpreting genetic variation and its consequences. Below are the types of mutations classified based on their nature and effects.
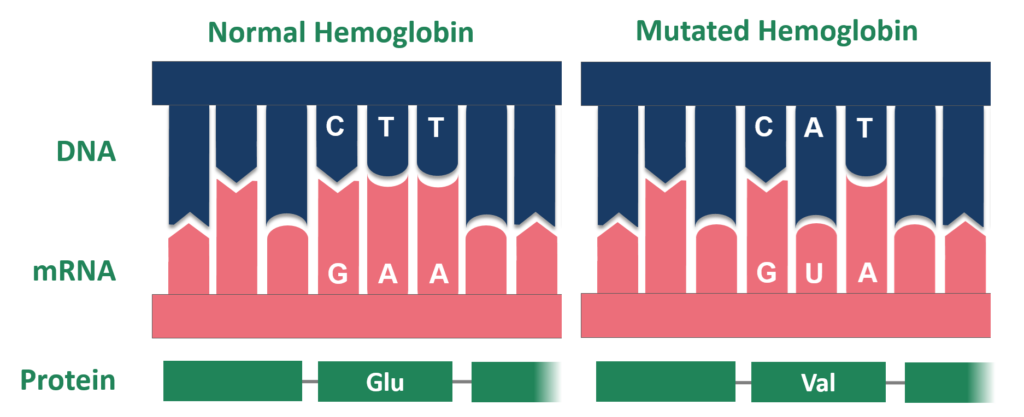
Point Mutations
Point mutations are changes involving a single nucleotide base pair. They are categorized into:
- Substitutions: A single base is replaced by a different base.
-
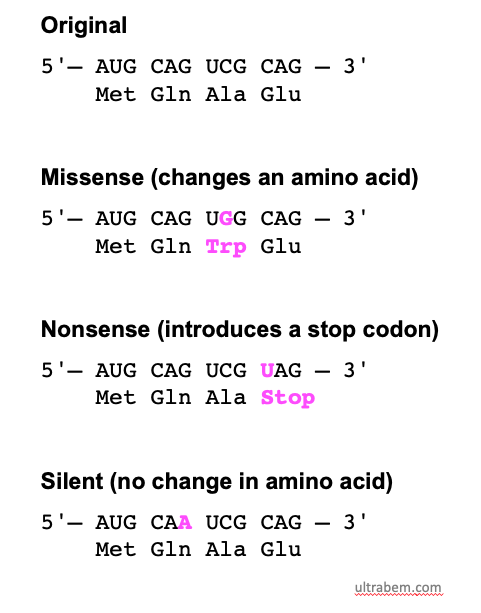
- Silent Mutation:
- The altered codon still codes for the same amino acid due to the redundancy of the genetic code.
- These mutations do not affect the protein and are often considered neutral.
- Missense Mutation:
- The altered codon codes for a different amino acid.
- This can result in a protein with altered function or no function, depending on the specific amino acid change.
- Nonsense Mutation:
- The altered codon becomes a stop codon, resulting in premature termination of translation.
- This typically produces a truncated protein that is often non-functional.
- Silent Mutation:
- Insertions and Deletions – Insertions and deletions involve the addition or removal of one or more nucleotide base pairs, which can lead to multiple consequences:
- Frameshift mutations:
- Insertion or deletion of a number of nucleotides that is not a multiple of three, shifting the reading frame of the gene.
- Frameshift mutations usually result in non-functional proteins because the entire downstream sequence is altered.
- In-frame mutations:
- Insertion or deletion of nucleotides in multiples of three, preserving the reading frame.
- These mutations may affect the function of the protein depending on the number and position of the inserted or deleted amino acids.
- Frameshift mutations:
- Structural Mutations – involve changes to larger sections of DNA, such as:
- Duplications – a segment of DNA is copied and inserted into the genome. Duplications can provide raw material for evolution, as the extra copy of a gene can accumulate mutations and potentially gain a new function.
- Deletions – segment of DNA is removed from the genome. Deletions can range from a few base pairs to large chromosomal regions, often resulting in loss of function.
- Inversions – a segment of DNA is reversed within the genome. Inversions typically do not affect gene function unless they disrupt regulatory regions or involve large segments.
- Translocations – a segment of DNA is moved to a different position within the genome. Translocations can result in fusion genes or disrupt regulatory elements, leading to altered gene expression.
Mutation by Impact on Function
Mutations can also be classified based on their impact on protein function. Loss-of-Function mutations result in a reduced or absent protein function and are often recessive because the presence of one functional copy of the gene can compensate for the loss. Gain-of-Function mutations result in a new or enhanced protein function. These are often dominant because the altered function can manifest even in the presence of a normal gene copy. Dominant-Negative Mutations are those where the mutated protein interferes with the function of the normal protein, often through abnormal interactions. These are typically dominant because they affect the function of both alleles.
Mutation by Origin
Lastly, mutations can be classified based on their origins. Germline mutations occur in the reproductive cells and are passed on to offspring. These mutations are important in evolution and hereditary disease. Somatic mutations occur in non-reproductive cells and are not passed on to offspring. These can lead to cancer and other somatic cell abnormalities.
THINK/PAIR/SHARE
- Consider the different outcomes of point mutations (silent, missense, and nonsense). How might each type of mutation influence the evolutionary fitness of an organism in a rapidly changing environment? Discuss scenarios where a missense or nonsense mutation could provide an adaptive advantage or disadvantage
- Given the descriptions of various mutation types, such as insertions, deletions, and structural mutations, discuss how these genetic alterations could lead to different genetic disorders or diseases. Can you think of specific examples where a frameshift mutation or a structural mutation has been identified as the cause of a particular disease? How do these mutations disrupt normal cellular function?
Mutational Rates
Mutational rates refer to the frequency at which mutations occur in a genome over a certain period of time or across a generation. Understanding mutation rates is crucial for elucidating the dynamics of genetic variation, which in turn impacts evolutionary processes, adaptation, and biodiversity. Mutation rates vary widely across species and even within genomes. Mutation rates tend to be lower in organisms with larger genomes, possibly due to the increased cost of correcting errors. In viruses, mutation rates are typically higher, which can facilitate rapid adaptation but also lead to higher deleterious mutation loads. Mutation rates also vary across different regions of the genome, often being higher in non-coding regions or areas with repetitive sequences The Y chromosome and mitochondrial DNA typically have higher mutation rates compared to autosomes, due to less effective repair mechanisms.
Table 1: Factors Affecting Mutational Rates
| Factor | Example | Explanation |
| Genetic Factors | 1. DNA Polymerase Fidelity
2. Mismatch Repair |
1. The accuracy of DNA polymerase during replication affects mutation rates.
2. Post-replication repair mechanisms correct mismatched based pairs, reducing mutation rates. |
| Environmental Factors | 1. Radiation
2. Chemicals 3. Temperature |
1. Exposure to UV or ionizing radiation increases mutation rates by damaging DNA.
2. Mutagenic chemicals can induce mutations. 3. High temperatures can increase mutation rates by destabilizing DNA. |
| Biological Factors | 1. Generation Time
2. Metabolic Rate |
1. Species with shorter generation times tend have higher mutation rates per year, as they undergo more rounds of replication.
2. Higher metabolic rates can increase mutation rates due to increased production of reactive oxygen species (ROS). |
Media Attributions
- Point-Mutation-Sickle-Cell-Normal_and_Mutated-Hemoglobin © Thomas Samuel - ACC Bioinnovation Lab is licensed under a CC BY-SA (Attribution ShareAlike) license
- DNA_mutation(1) is licensed under a CC0 (Creative Commons Zero) license

